Plants transform carbon dioxide from the environment into carbon-rich proteins in their tissues through a process known as carbon fixation. All of photosynthesis’ operations are directed toward this goal, and it is a critical component of the massive interconnected system that cycles carbon via plants, animals, microorganisms, and the atmosphere to keep life on Earth afloat.
Plants are not the true champions of carbon fixation as often thought, but soil microbes. Certain bacterial enzymes perform a vital stage in carbon fixation 20 times faster than plant enzymes. Understanding how they do this could help scientists construct artificial photosynthesis systems that transform greenhouse gases into fuels, fertilisers, medicines, and other useful things.
SLAC National Accelerator Laboratory, Stanford University, Germany’s Max Planck Institute for Terrestrial Microbiology, the DOE’s Joint Genome Institute (JGI), and Chile’s University of Concepción have all worked together to figure out how an enzyme, a molecular machine that helps with chemical reactions, gets going to do this job.
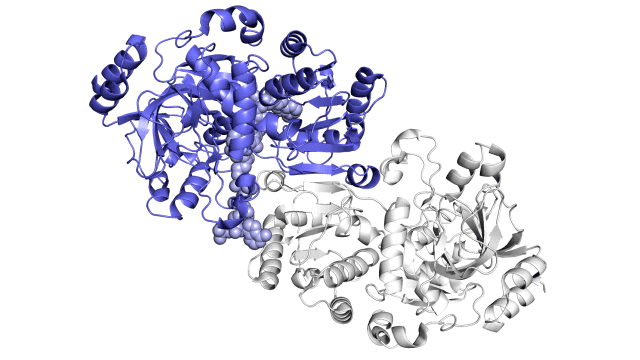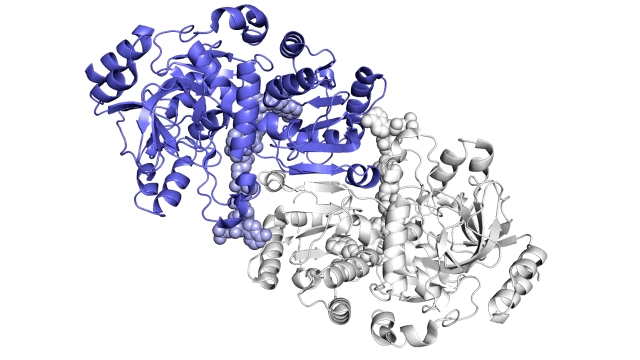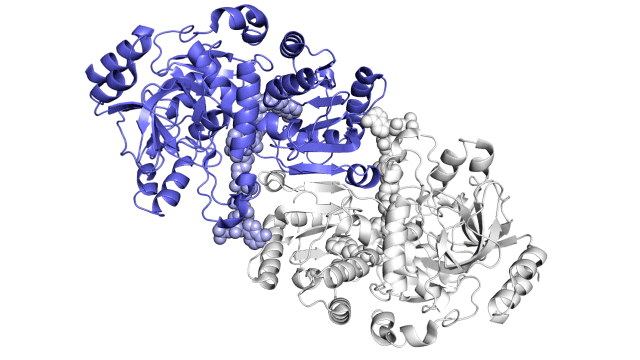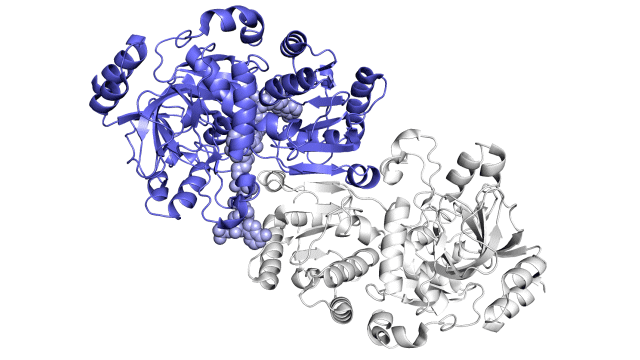
This enzyme is made up of pairs of molecules that act together in sync, similar to a juggler’s hands, which can throw and catch balls at the same time, rather than trapping carbon dioxide molecules and attaching them to proteins independently. A carbon-fixing enzyme pair is made up of two parts: one part that opens wide to get a specific set of reaction materials, and another part that shuts the materials in and performs the carbon-fixing reaction. The two parts then switch roles in a continuous process.
Researchers discovered that each pair of enzymatic hands is held together by a single molecular “glue,” allowing them to alternate opening and closing in a coordinated manner, while a twisting motion allows ingredients and finished products to be transported into and out of the pockets where reactions occur. The carbon-fixing reaction speeds up by a factor of a hundred thousand when both the glue and the twist are present.
One of the study’s researchers, SLAC and Stanford professor Soichi Wakatsuki explained that a multitude of enzymes in this family must work slowly but precisely to make a single product. Others are substantially faster and can produce chemical building blocks for a variety of products and materials. We can design enzymes that combine the benefits of both ways while operating extremely quickly with a wide range of starting materials now that we have better knowledge of the process.
enzyme story visuals 1
Nature has competition.
The enzyme in question belongs to the ECR (enoyl-CoA carboxylase/reductase) enzyme family. Enzymes that carboxylate and reduce enoyl-CoA are known as ECRs. It is made from Kitasatospora setae, a soil-dwelling bacteria capable of producing antibiotics as well as carbon fixation.
Tobias Erb of the Max Planck Institute for Terrestrial Microbiology in Germany and Yasuo Yoshikuni of the Japan Genome Institute originally taught Wakatsuki about this enzyme family half a dozen years ago (JGI). Erb’s research group has been working on bioreactors for artificial photosynthesis for a long time. These bioreactors can turn CO2 from the air into a variety of useful compounds.
Despite the fact that photosynthesis is necessary for life on Earth, it is inefficient, according to Erb. Every product made by millennia of evolution is only as good as it needs to be because it has been built on rather than made from scratch.
He further stressed that the stage of natural photosynthesis that involves the fixation of CO2 from the air, which is aided by an enzyme called Rubisco, is a bottleneck that slows down the entire chain of photosynthetic activities from start to finish. As a result, using enzymes that work quickly, as well as making them even faster, could make a big difference in productivity.
Erb made it clear that they were not attempting to replicate photosynthesis. They’re actually applying technical expertise to recreate natural principles in order to create a technique that’s significantly more efficient. Artificial chloroplasts, which are water droplets floating in oil, could be employed to produce this “photosynthesis 2.0” in both biological and synthetic systems.
Enzyme portraiture
Wakatsuki, his colleagues and the SLAC team had been researching a related process known as nitrogen fixation. This mechanism transforms nitrogen gas in the atmosphere into the chemicals that living organisms require. He began collaborating with Erb’s group in order to solve the mystery of why ECR enzymes could work so swiftly.
The SLAC researchers isolated and crystallised samples of the ECR enzyme in preparation for X-ray analysis at the Department of Energy’s Argonne National Laboratory’s Advanced Photon Source. All by itself and with a small helper molecule that helps it work, X-rays were used to figure out how the enzyme’s molecular structure looked.
In addition, X-ray studies at SLAC’s Stanford Synchrotron Radiation Lightsource (SSRL) revealed that when an enzyme was coupled to a substrate, the structure of the enzyme changed. A substrate is a molecular workbench that puts together and speeds up the parts needed for carbon fixation.
At the SACLA X-ray free electron laser in Japan, a team of researchers from the Linac Coherent Light Source (LCLS) at SLAC conducted further thorough examinations of the enzyme and its substrate. The researchers picked the X-ray laser because it allowed them to analyse the enzyme’s operation at room temperature, which is closer to the temperature of the enzyme’s natural environment, with little or no radiation damage.
To make sense of Wakatsuki’s structural discoveries, the Erb group in Germany and Associate Professor Esteban Vöhringer’s group at the University of Concepción in Chile conducted thorough biochemical research and dynamic simulations.
The simulations demonstrated that the opening and shutting of the enzyme’s two sections demands not only the use of molecular glue but also twisting motions around the central axis of each enzyme pair, according to Wakatsuki.
He says “this twist, almost like a rachet,” allows a finished product to be forced out or a new set of components to be brought into a pocket where the reaction occurs. When working together, the twisting and synchronisation of the enzyme pairs allows them to fix carbon 100 times per second.
A more versatile branch of the ECR enzyme family is capable of interacting with a wide range of biomolecules and producing a diverse range of products as a result of those interactions. Because they aren’t bonded together by molecular glue, their movements are uncoordinated, and they travel at a much slower rate than they would otherwise.
“If we can accelerate the rate of those challenging reactions that are used to synthesise novel proteins,” Wakatsuki says, “that would be a major advancement in the area.”
From static images to enzyme films.
So far, the research has shown static images of the enzyme, the parts of the reaction, and the finished products in a number of different ways. These images are shown below.
“Our dream experiment would be to combine all the ingredients as they flow into the path of the X-ray laser beam so we could watch the reaction take place in real time.,” Wakatsuki says.
He went on to say that the squad attempted to do so at SACLA but was unsuccessful. He says it’s impossible to catch the exact instant the CO2 molecules attach to the substrate because they’re so small and move so quickly. Because, among other things, the X-ray laser beam is too intense, we were unable to keep the components contained long enough for the reaction to occur.
He went on to suggest that an impending high-energy update to LCLS will likely fix the problem by transmitting pulses at a much higher rate – one million times per second – and allowing for individual control of sample power.
According to Wakatsuki, the LCLS sample delivery group is collaborating with researchers at the SLAC-Stanford cryo-EM facilities to figure out how to make this technology work.
Story Source: Original story written by Glennda Chui at DOE/SLAC National Accelerator Laboratory. Note: Content may be edited for style and length by Scible News.
Reference
Hasan DeMirci, Yashas Rao, Gabriele M. Stoffel, Bastian Vögeli, Kristina Schell, Aharon Gomez, Alexander Batyuk, Cornelius Gati, Raymond G. Sierra, Mark S. Hunter, E. Han Dao, Halil I. Ciftci, Brandon Hayes, Fredric Poitevin, Po-Nan Li, Manat Kaur, Kensuke Tono, David Adrian Saez, Samuel Deutsch, Yasuo Yoshikuni, Helmut Grubmüller, Tobias J. Erb, Esteban Vöhringer-Martinez, Soichi Wakatsuki. Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases. ACS Central Science, 2022; DOI: 10.1021/acscentsci.2c00057